In case of multicollinearity, there are columns (variables) in the data set, that are identical or linearly dependent. That happens, when columns are the exact multiple of another column. Our standard assumption of a \(\small{ N\times K }\) matrix is that \(\small{ rk\left(X\right)=K }\). However, when there is perfect multicollinearity, the rank of \(\small{ X }\) will be smaller than \(\small{ K }\).
In practice, perfect multicollinearity is quite rare. More often, we observe features that are nearly linearly dependent. That is called nearly perfect multicollinearity.
The problem of multicollinearity is that \(\small{ X^\prime X }\) is not regular (=singular) anymore. A singular matrix cannot be inverted. There is no solution for \(\small{ \left(X^\prime X\right)^{-1} }\) and hence, the OLS estimator \(\small{ \hat{\beta}={\left(X^\prime X\right)^{-1}\ X}^\prime y }\) is not identifiable.
A typical mistake, often seen among beginners, is the dummy trap. That is encoding a discrete variable with \(\small{ k }\) levels as \(\small{ k }\) individual dummy variables. There is always the case, that one level can be calculated by the other levels. Thus, the correct way of encoding is using \(\small{ k-1 }\) dummy variables.
Example: Correlation between Age and Years_since_schooling
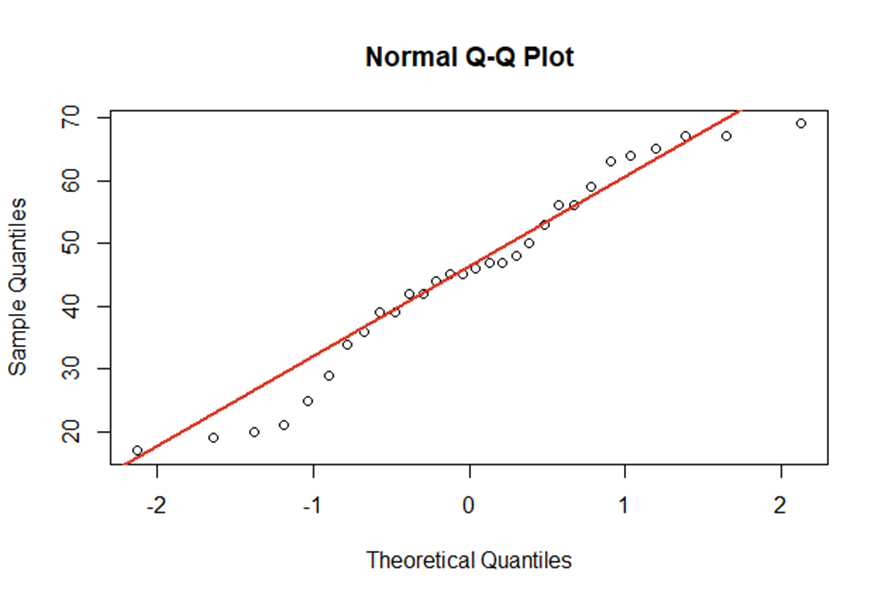
Multicollinearity can be identified by checking the correlation matrix of \(\small{ X }\) . The following example shows a high correlation between the age of a person and the years since graduation at school. Here, the result is quite intuitive.
| Age | Years_since_schooling | Blood_Pressure | |
| Age | 1.0000000 | 0.9899015 | 0.8439069 |
| Years_since_schooling | 0.9899015 | 1.0000000 | 0.8242502 |
| Blood_Pressure | 0.8439069 | 0.8242502 | 1.0000000 |
A strong correlation hints that the regressors are nearly perfect multicollinear. In result, the coefficients in regression analysis will not show high significance. They will be unstable and imprecise. Furthermore, we’d expect a high standard error rate. In some cases even numerical errors will arise by calculating the OLS regression.
Even though the coefficients are unstable and insignificant, the fitted values will be almost unchanged in many cases. The problem of nearly perfect multicollinearity hence lies in an improper understanding of the coefficients. The interpretation of the model will be extremely difficult.
Measurement for Multicollinearity
We already saw that the correlation between two variables can give us a hint about the presence of multicollinearity. Another common approach for identifying multicollinearity is the measurement of the variance inflation factor.
We define an auxiliary regression
\[{\hat{\beta}}_i=\left(x_i^\prime Q_{-i}x_i\right)^{-1}x_i^\prime Q_{-i}y\]
where \(\small{ Q_{-i}=I-P_{-i}=X_{-i}\left(X_{-i}^\prime X_{-i}\right)^{-1}X_{-i}^\prime }\) is the residual maker without the \(\small{ i }\)-th column \(\small{ x_i }\).
The coefficient of determination is hence
\[R_i^2=1-\frac{RSS_i}{S_{x_ix_i}}\]
where \(\small{ S_{x_ix_i} }\) is the variance of the \(\small{ i }\)-th column.
The variance inflation factor is now denoted as
\[VIF_i=\frac{1}{1-R_i^2} \]
As a rule of thumb, we’d expect no problem if \(\small{ VIF_i\le5 }\), but in case of \(\small{ VIF_i>10 }\) there seems to be serious multicollinearity. Then there are various possible options how to treat multicollinearity. The simplest way is ignoring it completely in those cases when the predictions are still valid and there seems to be no bad influence. If that is impossible, then one of the linear dependent variables can be omitted or transformed.